Frames Player and Analyzer for Optical Biological Ballistic Observations
by Luca Rogledi
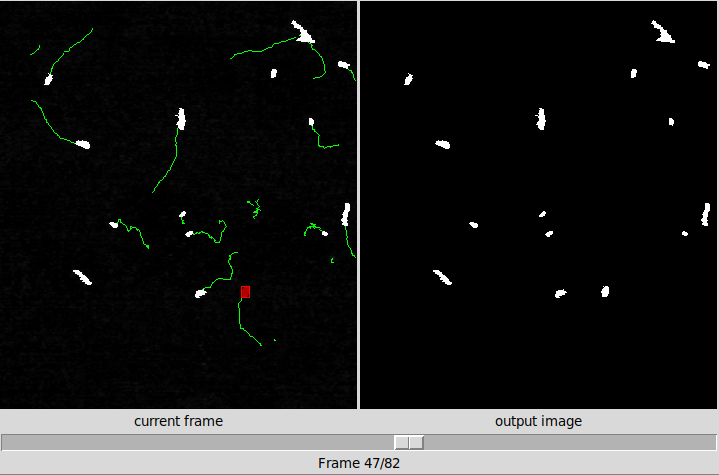
In the field of physiological research at the laboratory of Policlinico San Matteo of Pavia it was necessary to update the software used to assist the researchers in the study of the acto-myosin interaction. This is the work that led to the development of a program to perform such analysis.
Within this work three different approches have been explored in order to achieve this result: the Lucas Kanades optical flow, the linear assignment problem and the Kalman Filter.
Tools and Libraries
The application has been programmed with the Python language. The following tools and libraries have been used in this work:
- Numpy: Library that provides array and others data structures for optimized rutines and computations.
- pandas: Library that provides structures for data organization, useful for the export process.
- SciPy: Library that provides important algorithms in fields of engineering, math and science.
- openCV: Necessary open source library that includes computer vision algorithms.
- Matplotlibhttps://matplotlib.org/: Library for graphs representation, which was used to show the results.
- Pillow: Library that converts images in a tkinter compatible format.
- Tkinter: Library that allows the GUI implementation, through several widgets.
Goal of the Analysis
The main goal of the application is to track multiple actin filaments, showing the path of each cell within an implemented GUI. For this purpose, the user is allowed to set the parameters used for this process such as threshold, minimum size and maximum distance travelled by each cell.
The user must be able to observe paths and study the data of each cell within the path. The program elaborates useful statistics related to the motion of the filaments.
Application Description
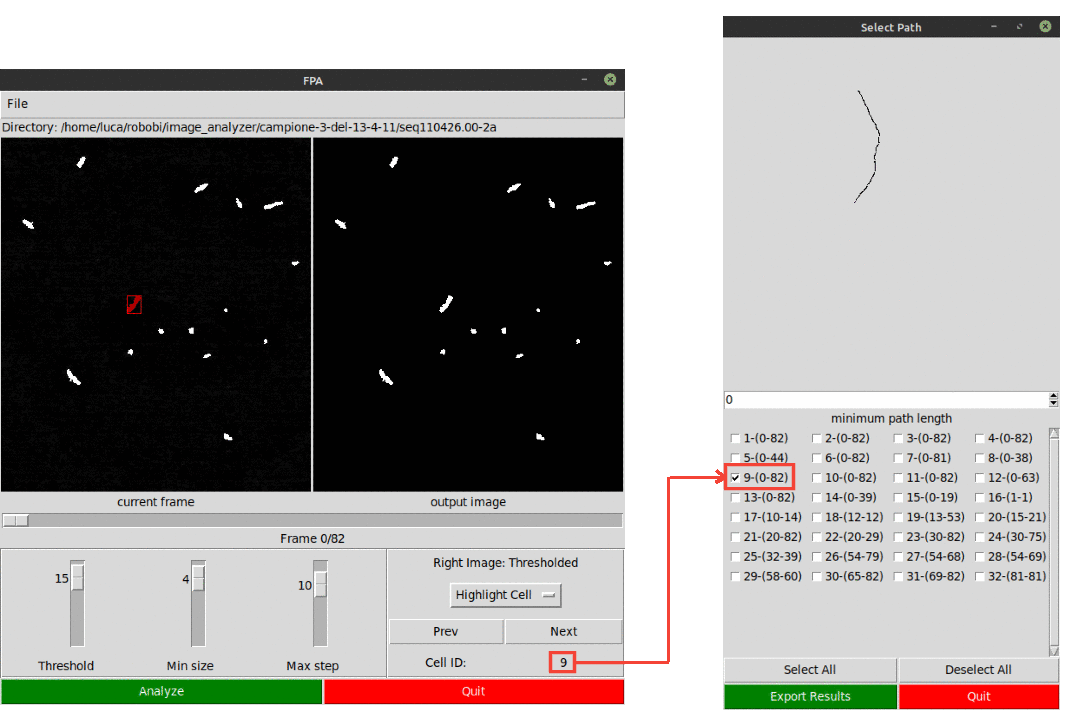
As can be seen in the picture, the GUI, which was created using Tkinter, contains the following “controls”:
- File menu
- Label directory
- Two canvases for image analysis
- Scale for frame selection
- Control panel, for set analysis parameters
- Display mode visualization menu
- Highlight control and information tools
- Analyze Function
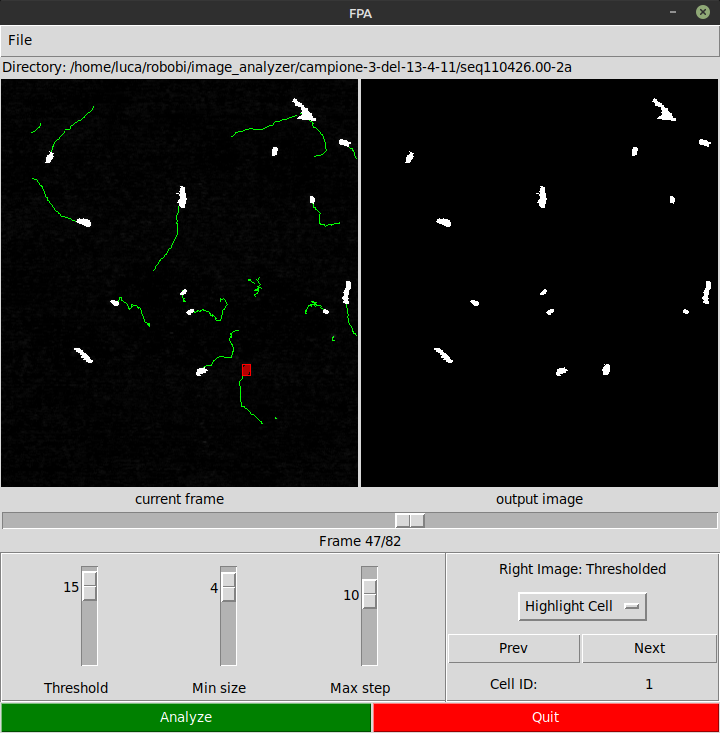
Application Features
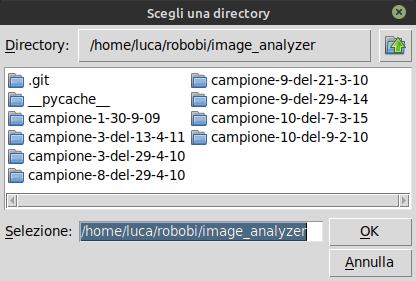
The application allows to perform the analysis by changing among different frames in the sequences of acquired frames. The set of frames composing an acquisition can be selectes with File menu option Select Directory.
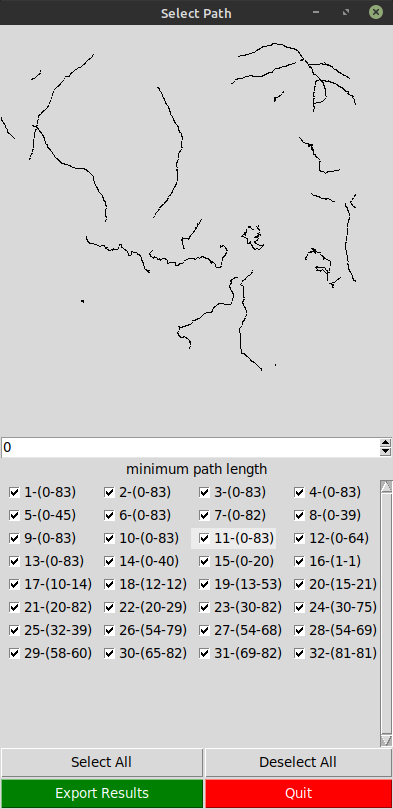
The user can select the desired paths to analyze using the File menu option Select Paths.
It is possible to highlight and identify a path using the Highlight Cell in the display menu options.
Results
The application creates a results directory for each analyzed sequence. The directory contains 6 files with the following purpose:
- 2 files with the data, in CSV format;
- 3 graphs showing the average values;
- 1 image representing the paths selected for the analysis.
The results.csv output file stores the data collected in each step of the sequence, including the x and y positions, x and y speeds and width and height of the actual bounding box of the cell.
The bounding box is the smallest rectangle that contains a cell’s contour and its information are expressed in pixel.
The x and y positions are expressed in micrometers [µm] and are necessary for the x and y speed computation in micrometers per second [µm/s].
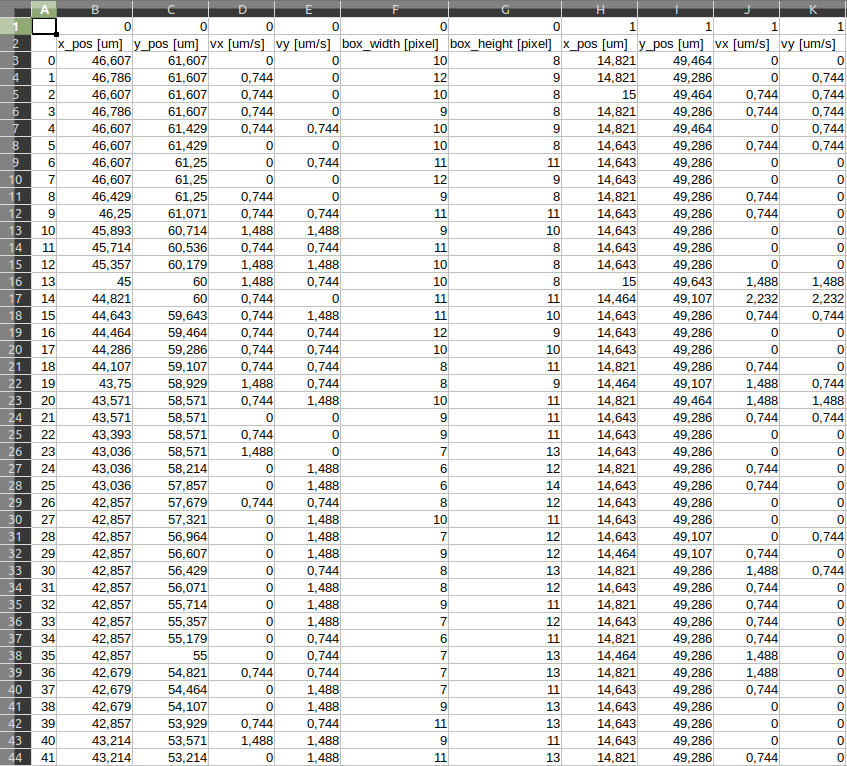
Similarly, the avg_results.csv contains information such as the speed-if-straight, which is the average speed of the path by supposing a strainght path, so considering only the first and the last position of the paths among all the frames.
This is useful to assess the “linearity” of the movement.
It also reports the avgerage speed of each path considering all changes in direction, the the length of the path, the first frame in which the path is detected in first-frame, and the initial x and y positions of the cell.
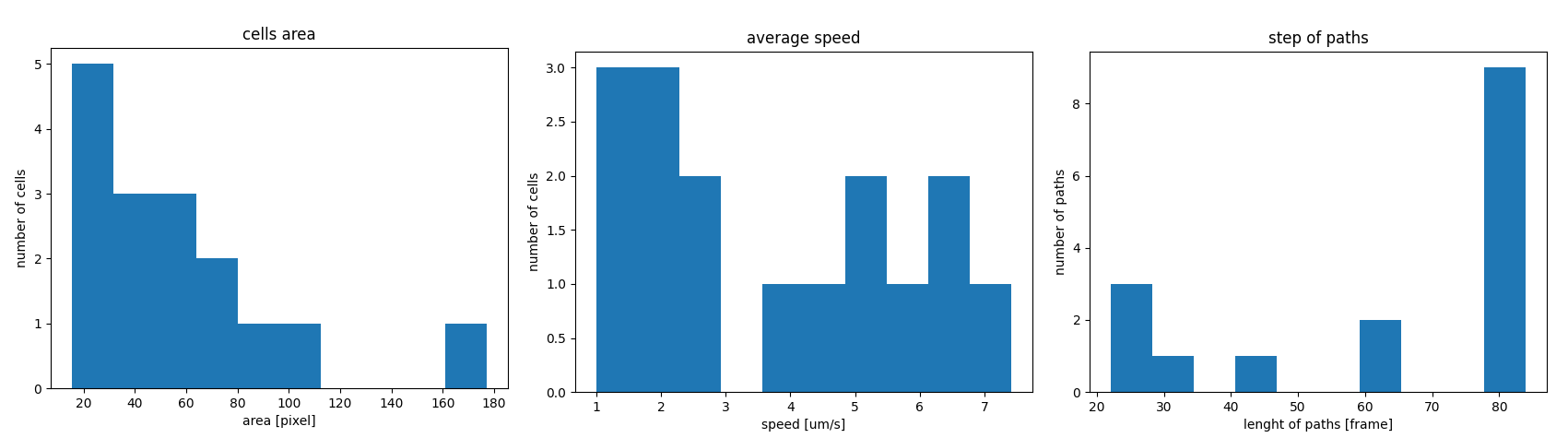
The graphs show the average area, speed and lenght data on the x axis and the number of related paths on the y axis. These graphs were a requirements of the reasearchers to allow them to get an immediate clue on the results of the acquisition.
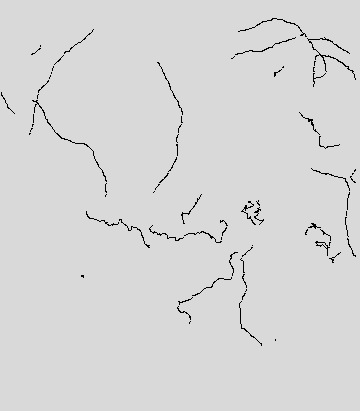
Finally, the image obtained from the selection of desired paths reports the paths selected for the data analysis using the Select Paths GUI.
Conclusions
For the tracking of the actin filaments was observed that the Lucas Kanade optical flow was not the best solution due to the good features to track not being tracked. For this reason, a tracking was implemented using the Kalman filter in order to predict the position of the cell and associate it minimizing the distance to the new detection.